CellOrganizer for Docker¶
About CellOrganizer for Docker¶
CellOrganizer for Docker is an image with compiled binaries from the CellOrganizer functions
img2slml, the top-level function to train generative models of cells, and
slml2img, the top-level function to generate simulated instances from a trained generative model.
slml2info, the top-level function to generate a report from information extracted from a single generative model.
slml2report, the top-level function to generate a report from comparing generative models.
slml2slml, the top-level function to combine models into a single model file.
The image also comes with
and the Python packages
ipywidgets 7.4
dill 0.2
dask 1.1
numba 0.42
bokeh 1.0
sqlalchemy 1.3
hdf5 1.10
vincent 0.4
protobuf 3.7
xlrd
About Docker¶
Docker performs operating-system-level virtualization. Docker lets us create and deploy a preconfigured image with the CellOrganizer binaries. This image can be used to spin containers that are ready to use and start testing.
To learn about Docker and how to use it, click here
About Jupyter¶
The Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations and narrative text.
To learn more about Jupyter, click here
Installing CellOrganizer for Docker¶
The instructions below describe
How to install Docker, the virtualization engine that will run the container
How to install Kitematic, a UI for Docker
How to download the latest cellorganizer-docker image from Docker Hub, i.e. the docker images repository
How to start a container from the Docker image
How to connect to the container
How to run some of the demos included in the container
Installing Docker¶
Before downloading the image and spinning a container, you need to install Docker. Installing Docker is beyond the scope of this document. To learn about Docker Community Edition (CE), click here.
Installing Kitematic and downloading CellOrganizer for Docker¶
The easiest way to download an image and run a container is to use Kitematic. Kitematic is a tool for downloading images and running containers.
To install Kitematic, click here.
Attention
Kitematic is not necessary, but it is recommended to streamline installation and deployment.
Download the most recent image using Kitematic
Start Kitematic. It should open a window similar to the screenshot below
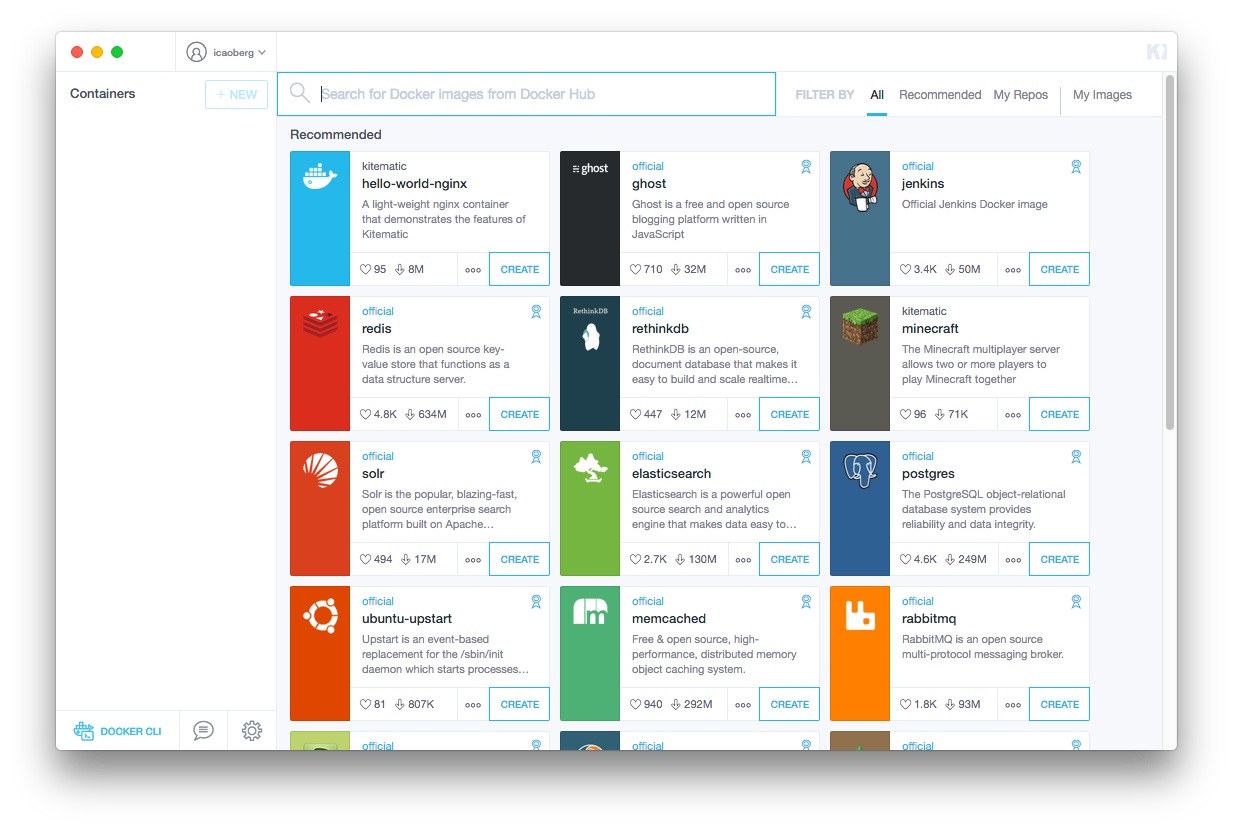
Searching for CellOrganizer should return a container like the image below
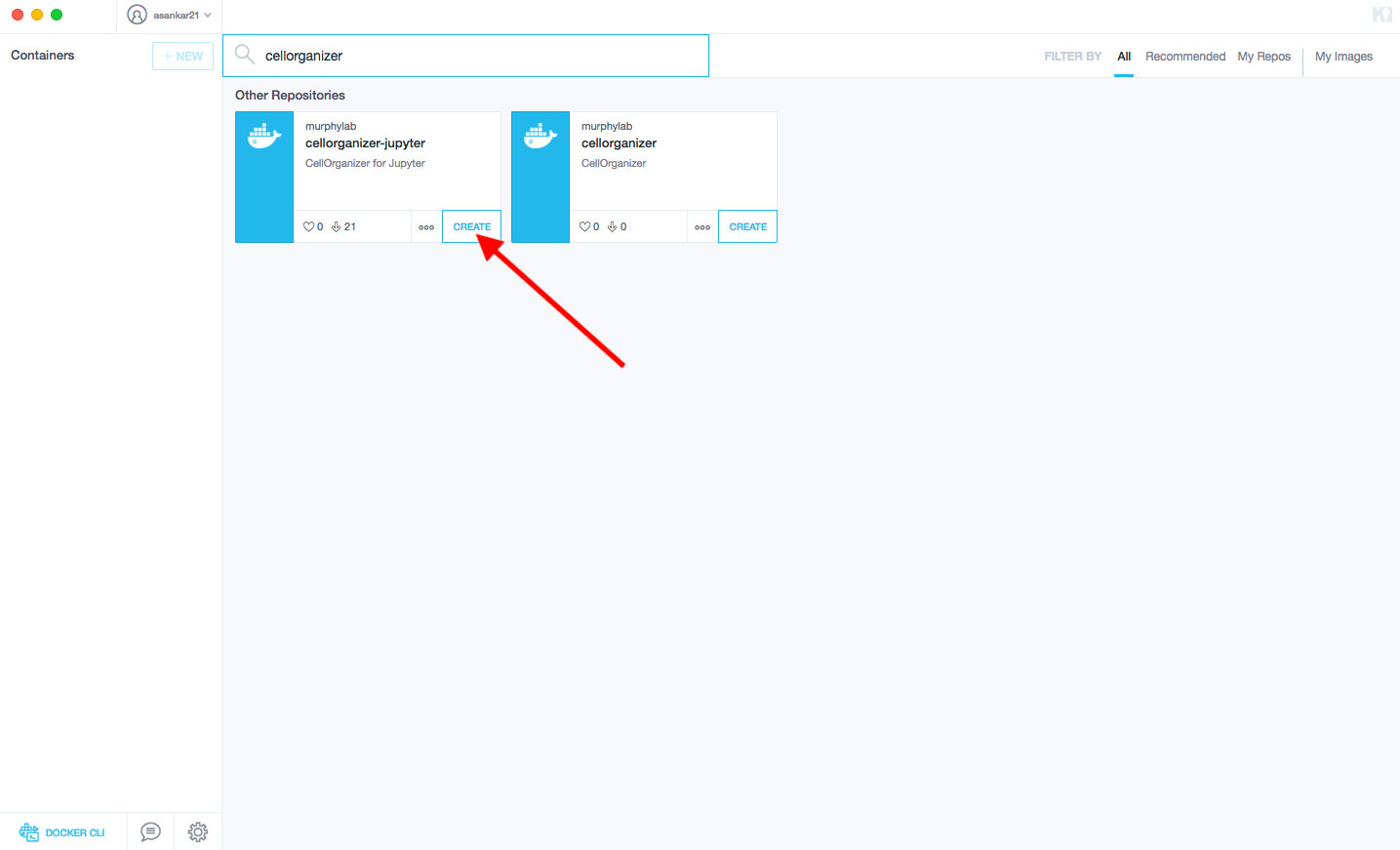
Then click CREATE to download the image and start a container
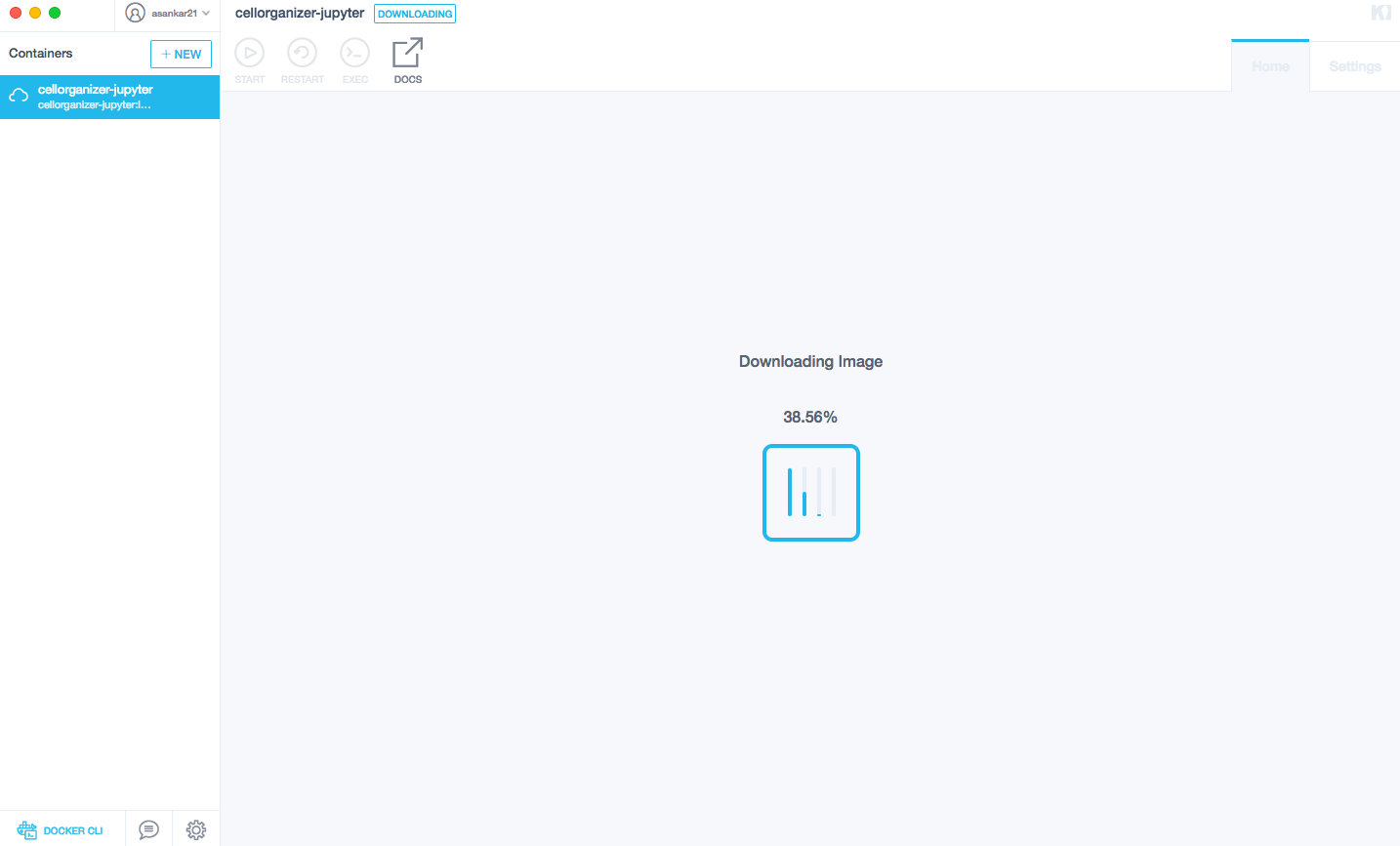
Download the most recent image from terminal
For convenience, the Docker image can be found in Docker Hub along with links to the Dockerfile.
Demos¶
There are several demos included within the CellOrganizer software bundle. These demos are intended to illustrate CellOrganizer’s functionality, and should be used to familiarize the user with the input/output format of various top-level functions such as img2slml and slml2img. Certains demos have been deprecated and will be removed in future versions of CellOrganizer.
Demo |
Training |
Synthesis |
Other |
Deprecated |
|---|---|---|---|---|
demo2D00 |
True |
|||
demo2D01 |
True |
|||
demo2D02 |
True |
|||
demo2D03 |
True |
X |
||
demo2D04 |
True |
|||
demo2D05 |
True |
|||
demo2D06 |
True |
|||
demo2D07 |
True |
|||
demo2D08 |
True |
|||
demo2D09 |
True |
|||
demo3D00 |
True |
|||
demo3D01 |
True |
|||
demo3D04 |
True |
|||
demo3D05 |
True |
|||
demo3D06 |
True |
X |
||
demo3D07 |
True |
|||
demo3D08 |
True |
|||
demo3D09 |
True |
|||
demo3D10 |
True |
|||
demo3D12 |
True |
|||
demo3D19 |
(True) |
Report |
||
demo3D25 |
True |
|||
demo3D47 |
Model |
|||
demo3D48 |
True |
|||
demo3D50 |
True |
|||
demo3D51 |
(True) |
Plot |
||
demo3D52 |
True |
|||
demo3D53 |
True |
|||
demo3D55 |
(True) |
Plot |
||
demo3D57 |
(True) |
Plot |
||
demo3D58 |
(True) |
|||
demo3D59 |
(True) |
Running CellOrganizer for Docker through Jupyter-Notebook¶
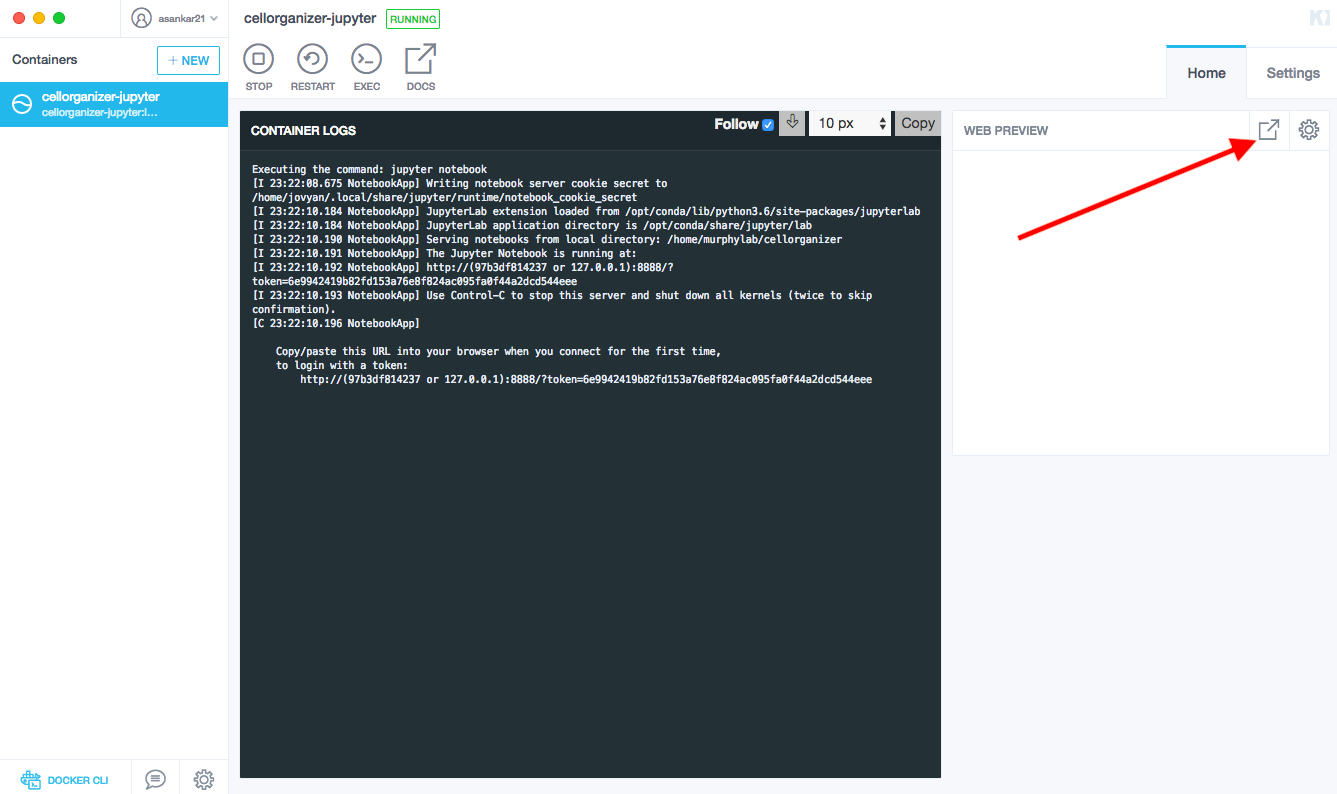
After the docker image has been created through Kitematic, click the top right button to open the Jupyter server.
This will open a webpage on your browser that is hosted by your local machine. The environment will intially contain Ipython notebooks with preinstalled demos that will run the CellOrganizer binaries.
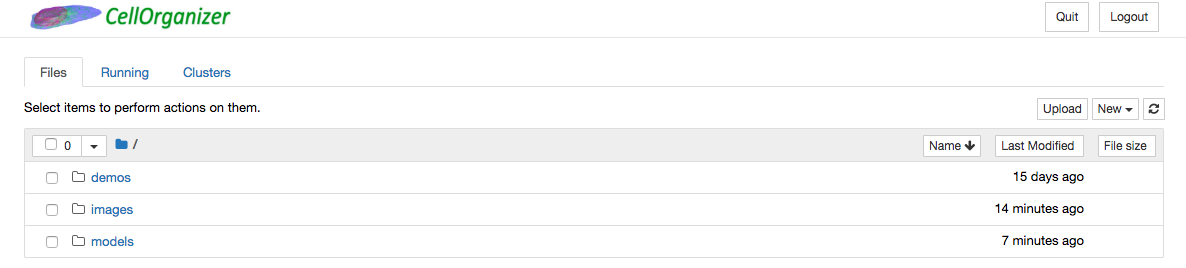
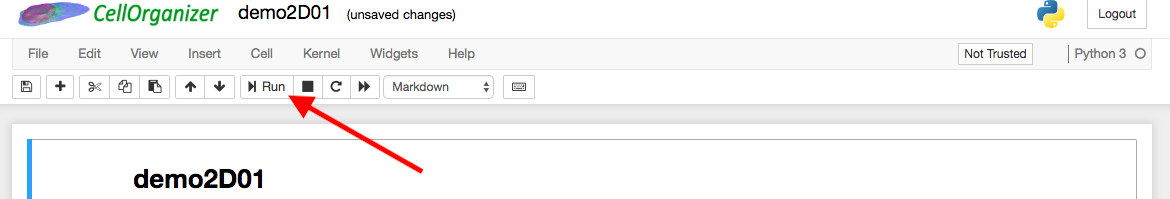
Run a demo that invokes img2slml¶
An example of a demo that trains a generative model from a series of .tif image files is demo2D01. To run a demo, simply click the run button at the top of the notebook.
This demo will save a folder param containing .mat files as well as a .mat file lamp2.mat to the same directory (/home/cellorganizer/demos/2D/demo2D01). These .mat files contain information characterizing the trained generative model.
Run a demo that invokes slml2img¶
An example of a demo that produces simulated images from a trained generative model is demo2D02.
This demo will save a folder img containing these simulated images to the same directory.
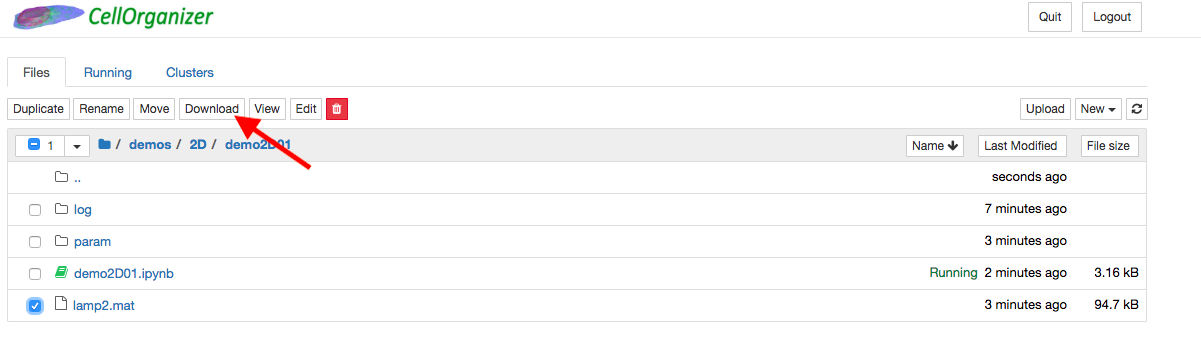
Export generated data out of the container¶
To export generated data out of the container, click the files in the directory that will be exported and click download.