Installation¶
Installing CellOrganizer locally¶
Requirements¶
- Matlab 2016b or newer
- Bioinformatics Toolbox
- Computer Vision System Toolbox
- Control System Toolbox
- Curve Fitting Toolbox
- Image Processing Toolbox
- Mapping Toolbox
- Optimization Toolbox
- Robust Control Toolbox
- Signal Processing Toolbox
- Simulink
- Simulink Design Optimization
- Statistics and Machine Learning Toolbox
- System Identification Toolbox
- Wavelet Toolbox
Downloading CellOrganizer¶
CellOrganizer for Matlab is the most flexible and powerful of the CellOrganizer deliverable, since it interfaces with Matlab which facilitates data analysis.
To download the latest CellOrganizer for Matlab distribution go to the download page. Aftwards, extract the contents of the release into a local directory of your preference.
For example,
cd ~/
wget -nc http://cellorganizer.org/downloads/v2.7/cellorganizer_v2.7.1_and_image_collection.tgz
tar -xvf cellorganizer_v2.7.1_and_image_collection.tgz
rm -fv cellorganizer_v2.7.1_and_image_collection.tgz
The commands above will download and extract to disk the contents of CellOrganizer v2.7.1.
Starting CellOrganizer¶
To start using CellOrganizer, start a Matlab session and change the directory to the location of CellOrganizer folder and run setup.m.
In the Matlab, type
>> cd( ‘/path/to/folder/cellorganizer’ );
>> setup();
If you were successful you should see a message like
>> setup
Checking for new stable version... Version is up to date.
You are now ready to use CellOrganizer for Matlab.
Installing CellOrganizer on a HPC¶
Requirements¶
Important
Remember to contact your HPC cluster managers to know if you have access to a Matlab license that would allow you to run Matlab in your cluster.
- Matlab 2016b or newer
- Bioinformatics Toolbox
- Computer Vision System Toolbox
- Control System Toolbox
- Curve Fitting Toolbox
- Image Processing Toolbox
- Mapping Toolbox
- Optimization Toolbox
- Robust Control Toolbox
- Signal Processing Toolbox
- Simulink
- Simulink Design Optimization
- Statistics and Machine Learning Toolbox
- System Identification Toolbox
- Wavelet Toolbox
Downloading CellOrganizer¶
Important
Sometimes, compute nodes do not access the web. Make sure to download the tarball from the front end or contact your HPC managers for further instructions.
CellOrganizer for Matlab is the most flexible and powerful of the CellOrganizer deliverable, since it interfaces with Matlab which facilitates data analysis.
To download the latest CellOrganizer for Matlab distribution go to the download page and download the latest release. Aftwards, extract the contents of the release into a local directory of your preference.
For example,
cd ~/
wget -nc http://cellorganizer.org/downloads/v2.7/cellorganizer_v2.7.1_and_image_collection.tgz
tar -xvf cellorganizer_v2.7.1_and_image_collection.tgz
rm -fv cellorganizer_v2.7.1_and_image_collection.tgz
The commands above will download and extract to disk the contents of CellOrganizer v2.7.1.
Starting CellOrganizer¶
The next instructions assume the HPC cluster you have access to uses SLURM as its default scheduler.
Important
Using CellOrganizer for Matlab on all possible popular schedulers is beyond the scope of this document, however feel free to contact us through the mailing list and we will do our best to help you.
Using CellOrganizer for Matlab interactively¶
Use srun or salloc to allocate resources to start Matlab.
For example
srun -p pool --mem=8Gb --pty /bin/bash
To start using CellOrganizer, start a Matlab session and change the directory to the location of CellOrganizer folder and run setup.m.
In the Matlab, type
cd( ‘/path/to/folder/cellorganizer’ );
setup();
If you were successful you should see a message like
>> setup
Checking for new stable version... Version is up to date.
You are now ready to use CellOrganizer for Matlab within that allocation or interactive session.
Submitting a job for CellOrganizer¶
Some tasks in CellOrganizer, with special attention on training, require considerable resources. At times, it will be more efficient to submit a job to a scheduler rather than doing it interactively.
For example, to run demo3D00, you could create a file called script.sh whose contents are
#!/bin/bash
#
#$ -j y
#$ -S /bin/bash
#$ -cwd
## the next line selects the partition/queue
#SBATCH -p pool
## the next line selects the number of cores
#SBATCH -n 4
## the next line selects the memory size
#SBATCH --mem=8G
## the next line selects the walltime
#SBATCH -t 00:30:00
cd /path/to/cellorganizer/folder
matlab -nodesktop -nosplash -r "setup(); demo3D00(), exit;"
Then use the command sbatch to submit it to the scheduler
sbatch script.sh
to add the job to the scheduler.
Installing CellOrganizer for Docker¶
Introduction¶
CellOrganizer for Docker contains binaries for the two main functions which this document will cover
- img2slml, the top-level function to train generative models of cell, and
- slml2img, the top-level function to generate simulated instances from a trained generative model.
- slml2info, the top-level function to generate a report from information extracted from a single generative model.
- slml2report, the top-level function to generate a report from comparing generative models.
Setup¶
The following instructions describe
- How to install Docker, the virtualization engine that will run the container
- How to install Kitematic, a UI for Docker
- How to download the latest cellorganizer-docker image from Docker Hub, i.e. the docker images repository
- How to start a container from the Docker image
- How to connect to the container
- How to run some of the demos included in the container
Source Code¶
For convenience, the Docker image can be found in Docker Hub along with links to the Dockerfile.
The instructions below will show you how to download and use this image.
Installing Docker¶
Before downloading the image and spinning a cotainer, you need to install Docker. Installing Docker is beyond the scope of this document. To learn about Docker Community Edition (CE), click here.
Download the most recent image using Docker command line
Open terminal and enter the command
docker pull murphylab/cellorganizer:latest
Running this command will initiate download and pull the most recent image of cellorganizer-docker from Docker Hub down to your computer.
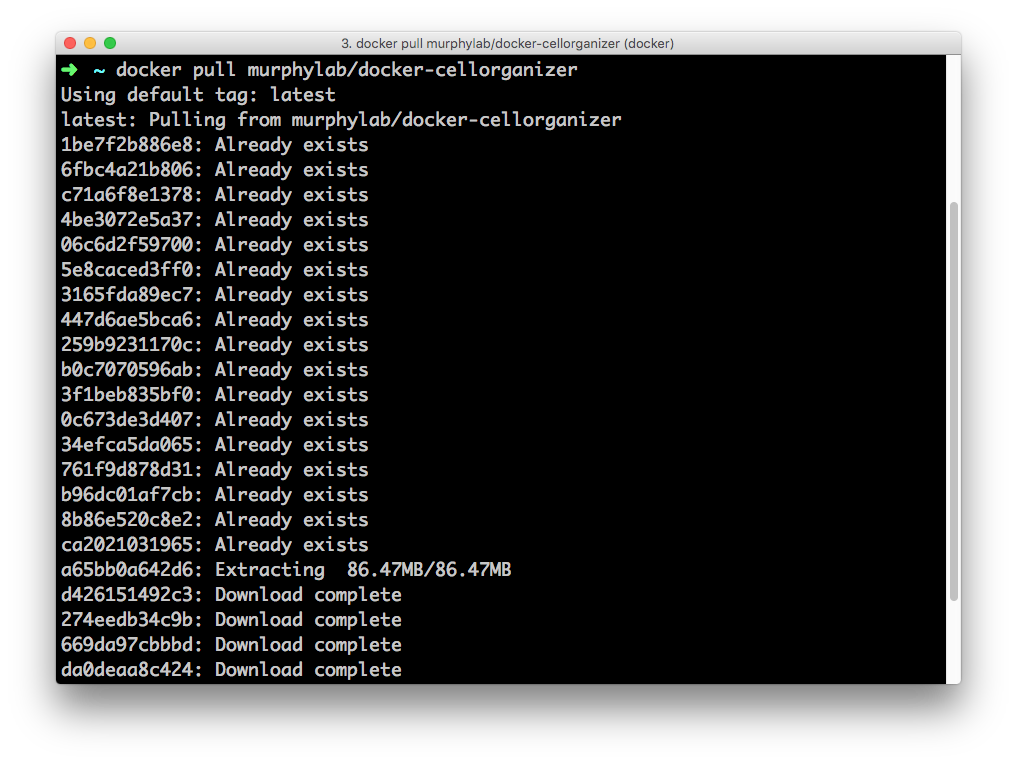
Once the download is complete, you can confirm the image was downloaded by entering the command:
docker images
You should see a record of a docker image identified by its repository murphylabs/cellorganizer and the tag latest.
Installing Kitematic¶
The easiest way to download an image and run a container is to use Kitematic. Kitematic is a tool for downloading images and running containers.
- To install Kitematic, click here.
Attention
Kitematic is not necessary but it is reccomended to streamline installation and deployment
Download the most recent image using Kitematic
Start Kitematic. It should open a window similar to the screenshot below
Demos included in the distribution¶
There are several demos included within the CellOrganizer software bundle. These demos are intended to illustrate CellOrganizer’s functionality, and should be used to familiarize the user with the input/output format of various top-level functions such as img2slml and slml2img.
