Demos¶
2D/3D Demos¶
For convenience, a series of demos are included with each distribution of CellOrganizer. These demos show
- how to synthesize images from existing models,
- how to train new models from raw data, as well as
- other functionality, e.g. exporting examples in multiple formats.
To display information about the available demos contained in the distribution, type in Matlab terminal:
>> demoinfo
Demos Summary Table¶
This table will let you know if the demo is meant to train a model or synthesize an image.
| Name | 2D/3D | Training | Synthesis | Diffeomorphic | Nucleus | Cell Shape | Protein pattern | Comment |
|---|---|---|---|---|---|---|---|---|
| demo2D00 | 2D | True | True | True | True | N/A | ||
| demo2D01 | 2D | True | True | True | True | N/A | ||
| demo2D02 | 2D | True | True | True | True | N/A | ||
| demo2D03 | 2D | True | True | True | True | N/A | ||
| demo2D04 | 2D | True | True | True | True | N/A | ||
| demo3D00 | 3D | True | True | True | True | N/A | ||
| demo3D01 | 3D | True | True | True | True | N/A | ||
| demo3D02 | 3D | surface plot | ||||||
| demo3D03 | 3D | True | True | True | True | N/A | ||
| demo3D04 | 3D | True | True | True | N/A | |||
| demo3D05 | 3D | True | True | True | True | N/A | ||
| demo3D06 | 3D | True | True | True | True | N/A | ||
| demo3D07 | 3D | True | True | True | True | N/A | ||
| demo3D08 | 3D | True | True | True | True | N/A | ||
| demo3D09 | 3D | True | True | True | True | N/A | ||
| demo3D10 | 3D | True | True | True | True | N/A | ||
| demo3D11 | 3D | True | True | True | True | N/A | ||
| demo3D12 | 3D | True | True | True | True | N/A | ||
| demo3D13 | 3D | syn2blender | ||||||
| demo3D14 | 3D | syn2projection | ||||||
| demo3D15 | 3D | True | True | True | True | True | N/A | |
| demo3D16 | 3D | preprocess raw images | ||||||
| demo3D18 | 3D | True | True | True | N/A | |||
| demo3D19 | 3D | slml2report | ||||||
| demo3D20 | 3D | True | True | True | True | N/A | ||
| demo3D21 | 3D | True | True | True | N/A | |||
| demo3D22 | 3D | True | True | True | True | True | N/A | |
| demo3D23 | 3D | True | True | True | True | N/A | ||
| demo3D24 | 3D | True | True | True | True | N/A | ||
| demo3D25 | 3D | True | True | True | True | SBML-Spatial format | ||
| demo3D26 | 3D | True | True | True | True | N/A | ||
| demo3D27 | 3D | True | True | True | time series | |||
| demo3D28 | 3D | SBML to SBML-Spatial | ||||||
| demo3D29 | 3D | True | N/A | N/A | ||||
| demo3D30 | 3D | True | True | N/A | N/A | |||
| demo3D31 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D32 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D33 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D34 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D35 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D36 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D37 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D38 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D39 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D40 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D41 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D42 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D43 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D44 | 3D | True | N/A | N/A | N/A | N/A | N/A | |
| demo3D45 | 3D | True | N/A | N/A | N/A | N/A | N/A |
Brief Descriptions¶
demo2D00¶
Demo header:
% Synthesize one 2D image with nuclear, cell shape, and vesicular channels
% from all vesicular object models (nucleoli, lysosomes, endosomes, and
% mitochondria) without convolution. The model was trained from the Murphy
% Lab 2D HeLa dataset.
%
% What you need
% -------------
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * one TIFF file with six slices (nuclear, cell shape, nucleolar,
% lysosomal, endosomal, and mitochondrial channels)
demo2D01¶
Demo header:
% Train 2D generative model of the nucleus, cell shape, and lysosome from
% all LAMP2 images in the Murphy Lab 2D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model file
demo2D02¶
Demo header:
% Synthesize one 2D image with nuclear, cell shape, and lysosomal channels
% from LAMP2 model trained in demo2D01 without convolution.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * one TIFF file with three slices (nuclear, cell shape, and lysosomal
% channels)
demo2D03¶
Demo header:
% Train 2D generative model of the nucleus, cell shape, and lysosome from
% all LAMP2 images in the Murphy Lab 2D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model file
demo2D04¶
Demo header:
% Train 2D generative diffeomorphic nuclear and cell shape model and a
% lysosomal model from all LAMP2 images in the Murphy Lab 2D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model file
demo3D00¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and nucleolar channels
% from nucleolar model with sampling method set to render nucleoli as
% ellipsoids without convolution. The model was trained from the Murphy Lab
% 3D HeLa dataset.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * three TIFF files (nuclear, cell shape, and nucleolar channels)
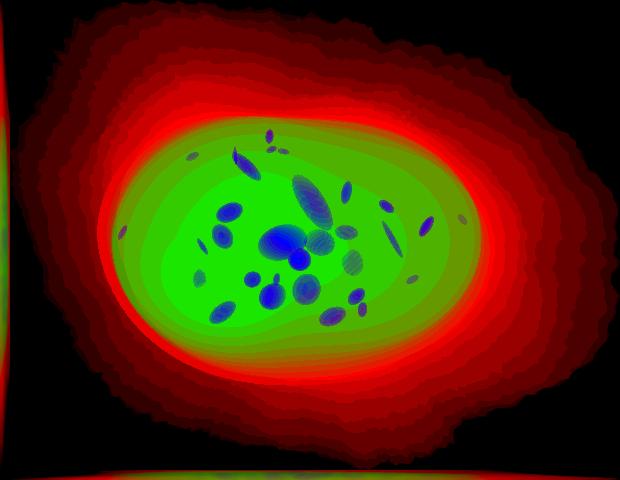
demo3D01¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and vesicular channels
% from all vesicular object models (lysosomes, mitochondria, nucleoli, and
% endosomes) with sampling method set to render vesicular objects as
% ellipsoids without convolution. The model was trained from the Murphy Lab
% 3D HeLa dataset.
%
% Input
% -----
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * six TIFF files (nuclear, cell shape, lysosomal, mitochondrial,
% nucleolar, and endosomal channels)
demo3D02¶
Demo header:
% Generate surface plot of image synthesized by demo3D00.
%
% Input
% -----
% * three TIFF files (nuclear, cell shape, and nucleolar channels)
% from demo3D00 directory
%
% Output
% ------
% * a surface plot of the synthetic image
demo3D03¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and vesicular channels
% from all vesicular object models (nucleoli, lysosomes, endosomes, and
% mitochondria) with sampling method set to sample vesicular objects from
% Gaussians at density 75 without convolution. The model was trained from
% the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * six TIFF files (nuclear, cell shape, nucleolar, lysosomal, endosomal,
% and mitochondrial channels)
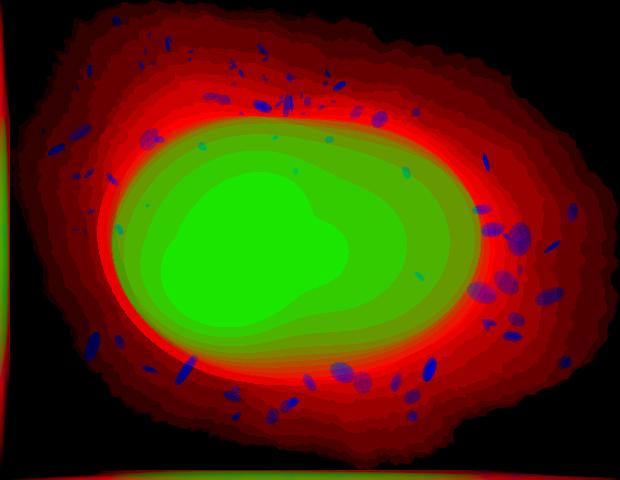
demo3D04¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and microtubule
% channels from microtubule model without convolution. The model was
% trained from the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a valid CellOrganizer centrosome model file
% * a valid CellOrganizer microtubule model file
%
% Output
% ------
% * three TIFF files (nuclear, cell shape, and microtubule channels)
demo3D05¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and protein channels
% from all object models (nucleoli, lysosomes, endosomes, mitochondria, and
% microtubules) with sampling method set to sample vesicular objects from
% Gaussians without convolution. The model was trained from the Murphy Lab
% 3D HeLa dataset.
%
% Input
% -----
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * seven TIFF files (nuclear, cell shape, nucleolar, lysosomal, endosomal,
% mitochondrial, and microtubule channels)
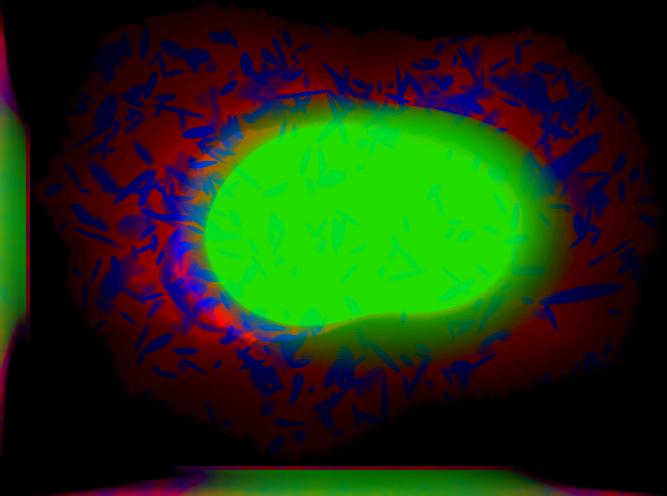
demo3D06¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and protein channels
% from all object models (nucleoli, lysosomes, endosomes, mitochondria, and
% microtubules) with sampling method set to render vesicular objects as
% ellipsoids and convolution with point-spread function. The model was
% trained from the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * seven TIFF files (nuclear, cell shape, nucleolar, lysosomal, endosomal,
% mitochondrial, and microtubule channels)
demo3D07¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and protein channels
% from all object models (nucleoli, lysosomes, endosomes, mitochondria, and
% microtubules) with sampling method set to sample vesicular objects from
% Gaussians at a density of 25 and convolution with point-spread function.
% The model was trained from the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * seven TIFF files (nuclear, cell shape, nucleolar, lysosomal, endosomal,
% mitochondrial, and microtubule channels)
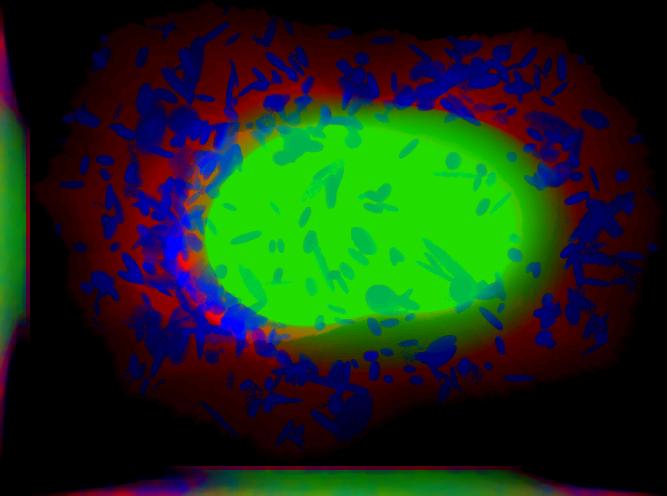
demo3D08¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and vesicular channels
% from all vesicular object models (nucleoli, lysosomes, endosomes, and
% mitochondria) with sampling method set to render vesicular objects as
% ellipsoids without convolution. The model was trained from the Murphy Lab
% 3D HeLa dataset.
%
% Input
% -----
% * a list of valid CellOrganizer model files
%
% Output
% ------
% * single indexed TIFF file which indexes the six TIFF files (nuclear,
% cell shape, nucleolar, lysosomal, endosomal, and mitochondrial channels)
demo3D09¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and lysosomal channels
% from LAMP2 model with sampling method set to render lysosomes as
% ellipsoids without convolution. Also render 2D mean projections along XY,
% XZ, and YZ axes of image. The model was trained from the Murphy Lab 3D
% HeLa dataset.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * three TIFF files (nuclear, cell shape, and lysosomal channels)
% * one projection TIFF file
% * one projection PNG file
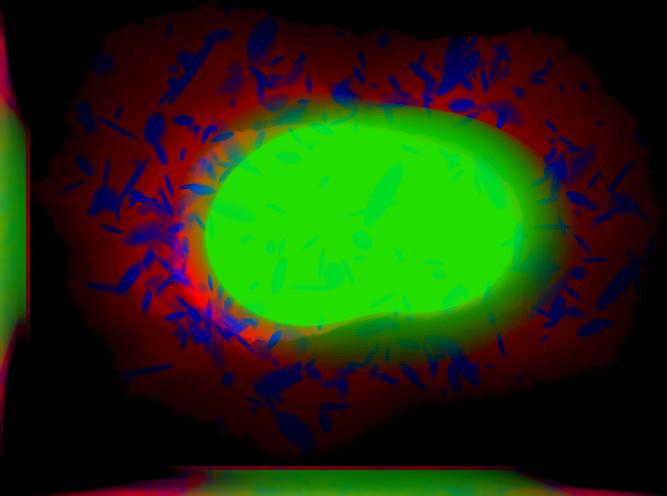
demo3D10¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and lysosomal channels
% with object files that can be imported to Blender from LAMP2 model,
% with sampling method set to render lysosomes as ellipsoids without
% convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * three TIFF files (nuclear, cell shape, and lysosomal channels)
% * three Wavefront OBJ files (nuclear, cell shape, and lysosomal channels)
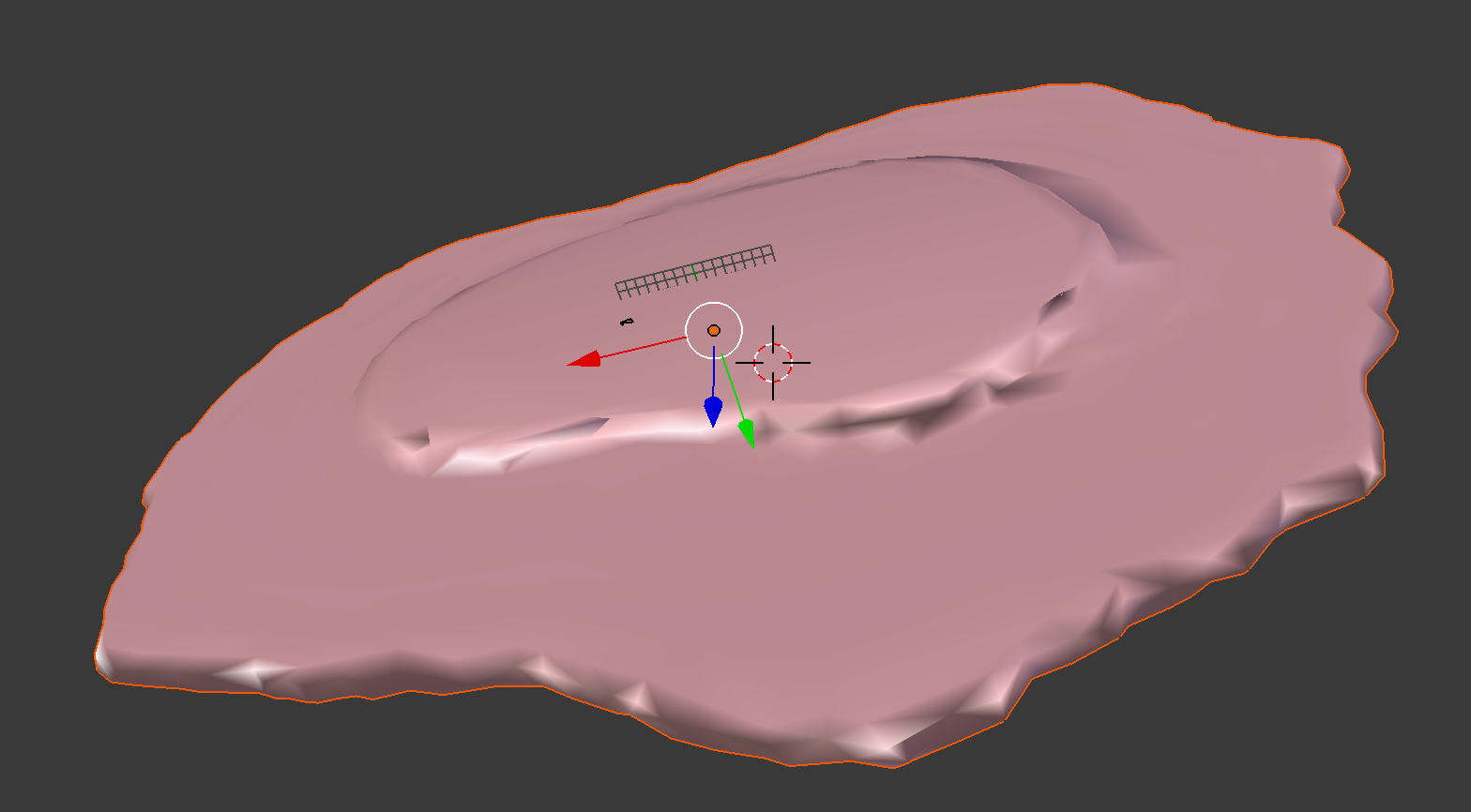
demo3D11¶
Demo header:
% Train 3D generative model of the cell framework (nucleus and cell shape)
% from the entire Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid model
demo3D12¶
Demo header:
% Train 3D generative model of the nucleus, cell shape, and lysosome from
% all LAMP2 images in the Murphy Lab 3D HeLa dataset that are either in the
% current directory or in the demo3D11 directory.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model file
demo3D13¶
Demo header:
% Export images synthesized by demo3D01 as object files importable to
% Blender.
%
% Input
% -----
% * a directory of 3D synthetic images
%
% Output
% ------
% * Wavefront OBJ files
demo3D14¶
Demo header:
% Render 2D mean projections along XY, XZ, and YZ axes of images
% synthesized by demo3D01.
%
% Input
% -----
% * a directory of 3D synthetic images
%
% Output
% ------
% * projections of synthetic images as TIFF files

demo3D15¶
Demo header:
% Synthesize one multichannel 3D image from an endosomal model and
% diffeomorphic nuclear and cell shape model. The sampling method was set
% to render endosomes as ellipsoids without convolution. The model was
% trained from the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a valid CellOrganizer model file with a diffeomorphic framework
%
% Output
% ------
% * three TIFF files (nuclear, cell shape, and endosomal channels)
demo3D16¶
Demo header:
% The main idea behind this demo is to show the user they
% can use their own binary images from raw experimental data
% to synthesize protein patterns. This demo uses the CellOrganizer
% method for nuclear and cell segmentation.
%
% The current demo assumes the resolution of the images is the same as
% the resolution of the images that were used to train the protein model.
%
% Input
% -----
% * raw or synthetic images of the nuclear and cell membrane
% * a valid CellOrganizer model file
%
% Output
% ------
% * three TIFF files (cell shape, nuclear, and lysosomal channels)
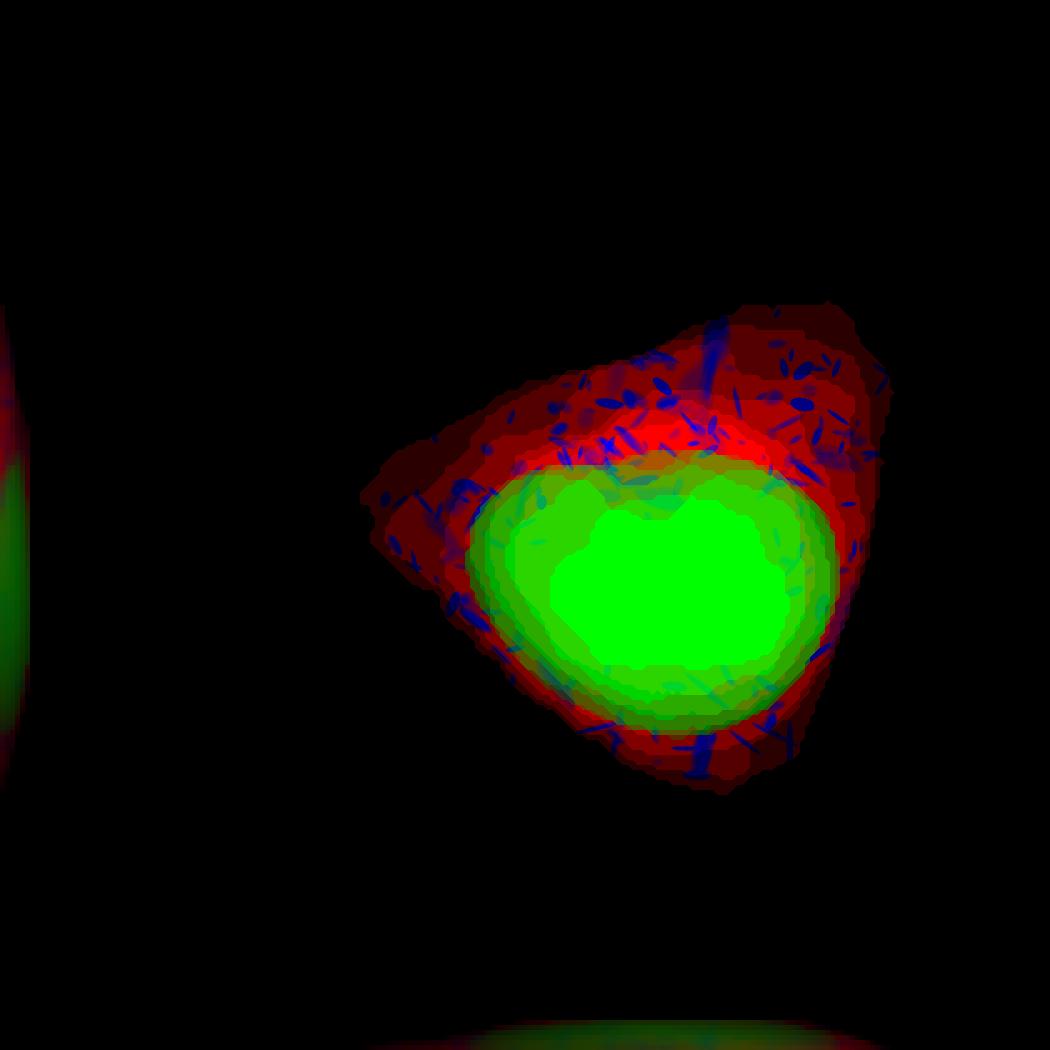
demo3D17¶
Demo header:
% The main idea behind this demo is to show the user they
% can use their own binary images from raw experimental data
% to synthesize protein patterns.
%
% The current demo assumes the resolution of the images is the same
% as the resolution of the images that were used to train the protein model.
%
% Input
% -----
% * an existing raw or synthetic framework, i.e. one binary multi-TIFF
% file of the nuclear channel and one binary multi-TIFF file of the
% cell membrane
% * the resolution of the latter images
% * a valid CellOrganizer model that contains a protein model
%
% Output
% ------
% * three TIFF files (cell shape, nuclear, and lysosomal channels)
demo3D18¶
Demo header:
% Train 3D generative model of the cell framework (nucleus and cell shape),
% using hole-finding to infer both nucleus and cell shape from the supplied
% protein pattern. The 3D 3T3 dataset was collected in collaboration with
% Dr. Jonathan Jarvik and Dr. Peter Berget.
%
% Input
% -----% * a directory of raw or synthetic protein images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model
demo3D19¶
Demo header:
% This demo uses slml2report to compare the parameters between
% CellOrganizer models and returns a report.
%
% Input
% -----
% * a set of valid CellOrganizer models
%
% Output
% ------
% * a report
demo3D20¶
Demo header:
% Train 3D generative diffeomorphic model of the cell framework (nucleus
% and cell shape) from all LAMP2 images in the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% -------
% * a valid SLML model file
% * a visualization of the shape space
demo3D21¶
Demo header:
% Train 3D generative model of the cell framework (nucleus and cell shape),
% using hole-finding to infer both nucleus and cell shape from the supplied
% protein pattern. This is identical to demo3D18 minus scaling the
% images. The 3D 3T3 dataset was collected in collaboration with Dr.
% Jonathan Jarvik and Peter Berget.
%
% Input
% -----
% * a directory of raw or synthetic protein images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model
demo3D22¶
Demo header:
% Synthesizes a protein pattern instance from the synthetic image produced
% in demo3DDiffeoSynth.
%
% Input
% -----
% * a synthetic framework
%
% Output
% ------
% * a synthetic image
demo3D23¶
Demo header:
% Train 3D generative diffeomorphic nuclear, cell shape, and a
% lysosomal model from all LAMP2 images in the Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model file
demo3D24¶
Demo header:
% This demo converts a sample SBML file to an SBML-spatial instance using
% the "matchSBML" function. This function takes an SBML file, matches the
% compartments in the file with available models and synthesizes the
% appropriate instances.
%
% Input
% -----
% * sample SBML file
%
% Output
% ------
% * valid SBML model
demo3D25¶
Demo header:
% Synthesizes 1 image using a lysosomal model with sampling mode
% set to 'disc', no convolution and output.SBML set to true.
% Results will be three TIFF files, one each for cell boundary,
% nuclear boundary, and lysosomes, in folder "synthesizedImages/cell1"
% Additionally, in the folder "synthesizedImages/" will be a
% SBML-Spatial(v0.82a) formatted .xml file containing constructed solid
% geometry(CSG) primitives for lysosomes and parametric objects for the
% cell and nuclear shapes.
%
% These files can then be read into VCell using the built in importer or
% CellBlender using the helper function provided in this distribution.
%
% Input
% -----
% * valid SBML model
%
% Output
% ------
% * three TIFF files
% * XML file with primitives for lysosomes and parametric objects
demo3D26¶
Demo header:
% This function displays a shape space of some dimensionality. This demo
% uses the model trained in Johnson 2015.
%
% Input
% -----
% * a CellOrganizer diffeomorphic model
%
% Output
% ------
% * a display of the shape space
demo3D27¶
Demo header:
% This demo performs a regression between two sets of related shapes (i.e.
% predicts cell shape from nuclear shape) and displays the residuals as in
% Figure 2 of Johnson et al 2015.
%
% Input
% -----
% * models hela_cell_10_15_15.mat and hela_nuc_10_15_15.mat
%
% Output
% ------
% * shape space figure
demo3D28¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape, and nucleolar channels
% from nucleolar model with sampling method set to render nucleoli as
% ellipsoids without convolution. The model was trained from the Murphy Lab
% 3D HeLa dataset.
%
% Input
% -----
% * an existing raw or synthetic nuclear image, i.e. one binary multi-TIFF
% file of the nuclear channel
% * the resolution of the input image
% * a valid CellOrganizer model that contains a cell membrane model
%
% Output
% ------
% * three TIFF files (cell shape, nuclear, and nucleolar channels)
demo3D29¶
Demo header:
% This demo illustrates different ways to sample from points in a
% diffeomorphic model.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * a random walk
demo3D30¶
Demo header:
% This demo illustrates how to sample uniformly at random from a
% diffeomorphic model.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * a random walk
demo3D32¶
Demo header:
% Synthesizes 1 image using a lysosomal model with sampling mode
% set to 'disc', no convolution using the object avoidance methods
% Results will be three TIFF files, one each for cell boundary,
% nuclear boundary, and lysosomes, in folder "synthesizedImages/cell1".
%
% Input
% -----
% * valid SBML file
%
% Output
% ------
% * three TIFF files
demo3D33¶
Demo header:
% Synthesize multiple 3D images from a lysosome model, at different resolutions.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% -------
% * multiple instances of the same cell at different resolutions
demo3D34¶
Demo header:
% Synthesize one 3D image with nuclear, cell shape and a vesicular channel.
% This demo exports the synthetic image as an OME.TIFF as well as an
% SBML Spatial instance.
%
% Input
% -----
% * a valid CellOrganizer model
%
% Output
% ------
% * OME.TIFF
% * SBML instance
% * single channel TIF files
demo3D35¶
Demo header:
% Learns a random walk from time series images as in Figure 6 and 7 of
% Johnson 2015
%
% What you need
% -------------
% * a valid CellOrganizer diffeomorphic model
%
% Output
% ------
% * a viz of the shape space
demo3D36¶
Demo header:
% Synthesize multiple 3D images from a lysosome model at different resolutions.
%
% Input
% -----
% * valid lysosomal model
%
% Output
% ------
% * multiple 3D images at different resolutions
demo3D37¶
Demo header:
% This demo exists to illustrate how padding size and window size effect the
% performance of diffeomorphic metric.
demo3D38¶
Demo header:
% Synthesizes 1 image using a lysosomal model with sampling mode
% set to 'disc', no convolution using the object avoidance methods
% Results will be three TIFF files, one each for cell boundary,
% nuclear boundary, and lysosomes, in folder "synthesizedImages/cell1".
demo3D39¶
Demo header:
% This demo illustrates how to sample uniformly at random from a
% diffeomorphic model.
%
% Input
% -----
% * a valid CellOrganizer model file
%
% Output
% ------
% * a random walk
demo3D40¶
Demo header:
% Train 3D generative model of the cell framework (nucleus and cell shape)
% from the entire Murphy Lab 3D HeLa dataset.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid model
demo3D41¶
Demo header:
% Train 3D generative model of the nucleus, cell shape, and lysosome from
% all LAMP2 images in the Murphy Lab 3D HeLa dataset that are either in the
% current directory or in the demo3D11 directory.
%
% Input
% -----
% * a directory of raw or synthetic nucleus images
% * a directory of raw or synthetic cell shape images
% * a directory of raw or synthetic lysosome images
% * the resolution of the images (all images should have the same
% resolution)
%
% Output
% ------
% * a valid SLML model file
demo3D42¶
Demo header:
% this demo illustrates using CellOrganizer to train a protein distribution
% model following the approach described in
%
% K. T. Roybal, T. E. Buck, X. Ruan, B. H. Cho, D. J. Clark, R. Ambler,
% H. M. Tunbridge, J. Zhang, P. Verkade, C. Wülfing, and R. F. Murphy (2016)
% Computational spatiotemporal analysis identifies WAVE2 and Cofilin as
% joint regulators of costimulation-mediated T cell actin dynamics.
% Science Signaling 9:rs3. doi: 10.1126/scisignal.aad4149.
%
% The slowest step, which typically takes about 1 min per cell per frame,
% is to align each cell at each time to the standardized template.
% This demo uses 46 cells so it will take about 1 hour on a single core.
%
% Input
% -----
% * image and annotation files for one or more proteins for one or more
% time points
% > the default is to use images from the paper of LAT at time 0 - downloading the
% needed images requires about 4 GB of free disk space
%
% Output
% ------
% * a model for the average concentration in each voxel of a standardized
% cell shape (in demos/LAT_reltime_1.mat)
% * various intermediate results files (in /param and /tmp)
demo3D43¶
Demo header:
% This is the synthesis demo for T cell model.
% The demo takes in two models: one model contains both cell and nuclear
% shape models, and the other contains a T cell protein shape model. Same
% as other synthesis framework, it calls slml2img for the synthesis. The
% meanings of the options are commented in the script.
%
% Input
% -----
% * A protein model with type standardized map halp-elipsoid
% * A framework model the provide the shape of the cell.
%
% Output
% ------
% * one or more set(s) of synthesized images with cell shape and protein
% pattern.
