Demos¶
2D/3D Demos¶
For convenience, a series of demos are included with each distribution of CellOrganizer. These demos show
- how to synthesize images from existing models,
- how to train new models from raw data, as well as
- other functionality, e.g. exporting examples in multiple formats.
To display information about the available demos contained in the distribution, type in Matlab terminal:
>> demoinfo
Demos Summary Table¶
This table will let you know if the demo is meant to train a model or synthesize an image.
| Name | 2D/3D | Training | Synthesis | TIFF | Blender | SBML |
|---|---|---|---|---|---|---|
| demo2D00 | 2D | True | True | |||
| demo2D01 | 2D | True | ||||
| demo2D02 | 2D | True | True | |||
| demo2D03 | 2D | True | True | |||
| demo2D04 | 2D | True | True | |||
| demo3D00 | 3D | True | True | |||
| demo3D01 | 3D | True | True | |||
| demo3D02 | 3D | True | ||||
| demo3D03 | 3D | True | True | |||
| demo3D04 | 3D | True | True | |||
| demo3D05 | 3D | True | True | |||
| demo3D06 | 3D | True | True | |||
| demo3D07 | 3D | True | True | |||
| demo3D08 | 3D | True | True | |||
| demo3D09 | 3D | True | True | |||
| demo3D10 | 3D | True | True | |||
| demo3D11 | 3D | True | ||||
| demo3D12 | 3D | True | ||||
| demo3D13 | 3D | True | ||||
| demo3D14 | 3D | True | ||||
| demo3D15 | 3D | True | True | |||
| demo3D16 | 3D | True | ||||
| demo3D18 | 3D | True | True | |||
| demo3D19 | 3D | True | ||||
| demo3D20 | 3D | True | True | |||
| demo3D21 | 3D | True | True | |||
| demo3D22 | 3D | True | True | True | ||
| demo3DMultiresSynth | 3D | True | True | |||
| demo3DObjectAvoidance | 3D | True | True | |||
| demo3DPrimitives | 3D | True | True | True | True | |
| demo3DDiffeoSynth | 3D | True | True | |||
| demo3DDynamic | 3D | True | ||||
| demo3DSBML | 3D | True | True | |||
| demo3Dimg2sbml | 3D | True | True | True |
Brief Descriptions¶
demo2D00¶
Synthesize one 2D image with nuclear, cell shape, and vesicular channels from all vesicular object models (nucleoli, lysosomes, endosomes, and mitochondria) without convolution. The model was trained from the Murphy Lab 2D HeLa dataset.
demo2D01¶
Train 2D generative model of the nucleus, cell shape, and lysosome from all LAMP2 images in the Murphy Lab 2D HeLa dataset.
demo2D02¶
Synthesize one 2D image with nuclear, cell shape, and lysosomal channels from LAMP2 model trained in demo2D01 without convolution.
demo2D03¶
Train 2D generative model of the nucleus, cell shape, and lysosome from all LAMP2 images in the Murphy Lab 2D HeLa dataset.
demo2D04¶
Train 2D generative diffeomorphic nuclear and cell shape model and a lysosomal model from all LAMP2 images in the Murphy Lab 2D HeLa dataset.
demo3D00¶
Synthesize one 3D image with nuclear, cell shape, and nucleolar channels from nucleolar model with sampling method set to render nucleoli as ellipsoids without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
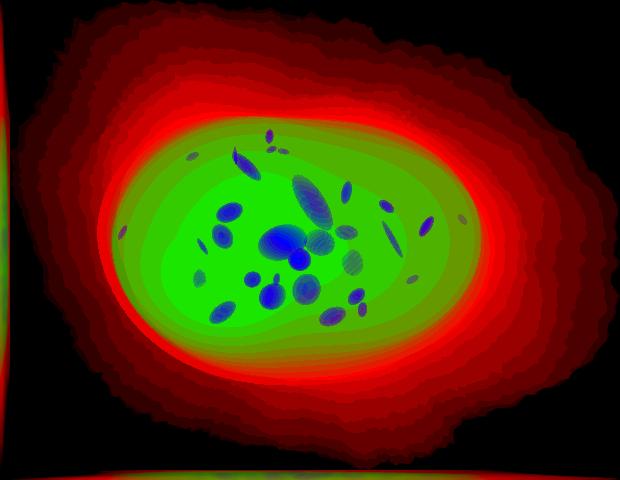
demo3D01¶
Synthesize one 3D image with nuclear, cell shape, and vesicular channels from all vesicular object models (lysosomes, mitochondria, nucleoli, and endosomes) with sampling method set to render vesicular objects as ellipsoids without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
demo3D03¶
Synthesize one 3D image with nuclear, cell shape, and vesicular channels from all vesicular object models (nucleoli, lysosomes, endosomes, and mitochondria) with sampling method set to sample vesicular objects from Gaussians at density 75 without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
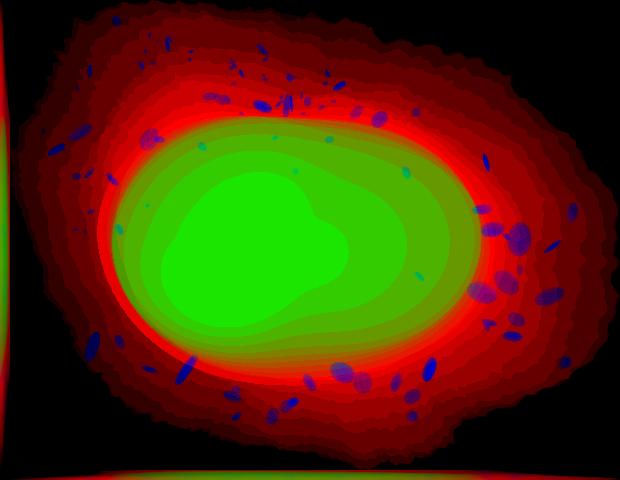
demo3D04¶
Synthesize one 3D image with nuclear, cell shape, and microtubule channels from microtubule model without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
demo3D05¶
Synthesize one 3D image with nuclear, cell shape, and protein channels from all object models (nucleoli, lysosomes, endosomes, mitochondria, and microtubules) with sampling method set to sample vesicular objects from Gaussians without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
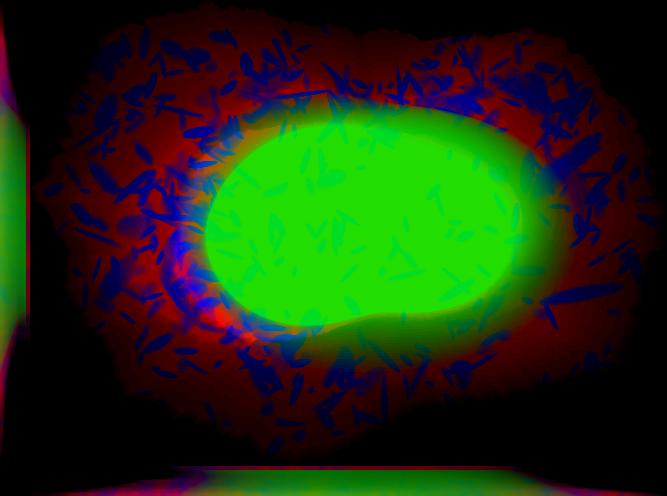
demo3D06¶
Synthesize one 3D image with nuclear, cell shape, and protein channels from all object models (nucleoli, lysosomes, endosomes, mitochondria, and microtubules) with sampling method set to render vesicular objects as ellipsoids and convolution with point-spread function. The model was trained from the Murphy Lab 3D HeLa dataset.
demo3D07¶
Synthesize one 3D image with nuclear, cell shape, and protein channels from all object models (nucleoli, lysosomes, endosomes, mitochondria, and microtubules) with sampling method set to sample vesicular objects from Gaussians at a density of 25 and convolution with point-spread function. The model was trained from the Murphy Lab 3D HeLa dataset.
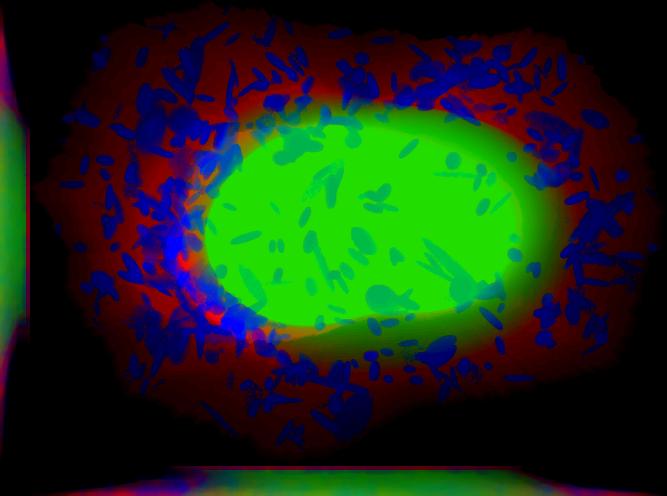
demo3D08¶
Synthesize one 3D image with nuclear, cell shape, and vesicular channels from all vesicular object models (nucleoli, lysosomes, endosomes, and mitochondria) with sampling method set to render vesicular objects as ellipsoids without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
demo3D09¶
Synthesize one 3D image with nuclear, cell shape, and lysosomal channels from LAMP2 model with sampling method set to render lysosomes as ellipsoids without convolution. Also ender 2D mean projections along XY, XZ, and YZ axes of image. The model was trained from the Murphy Lab 3D HeLa dataset.
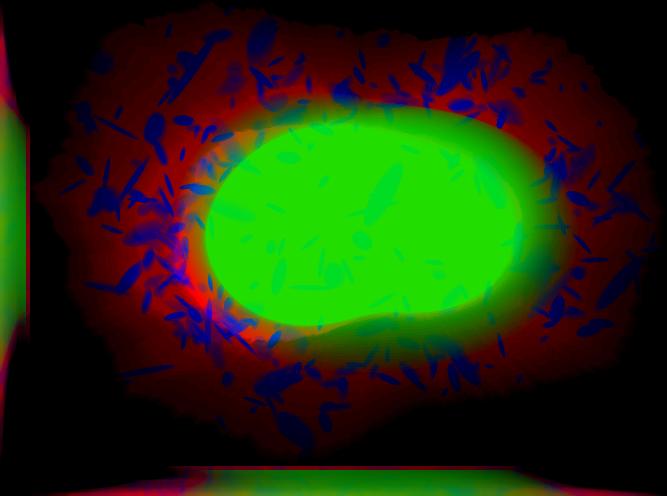
demo3D10¶
Synthesize one 3D image with nuclear, cell shape, and lysosomal channels with object files importable to Blender from LAMP2 model, with sampling method set to render lysosomes as ellipsoids without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
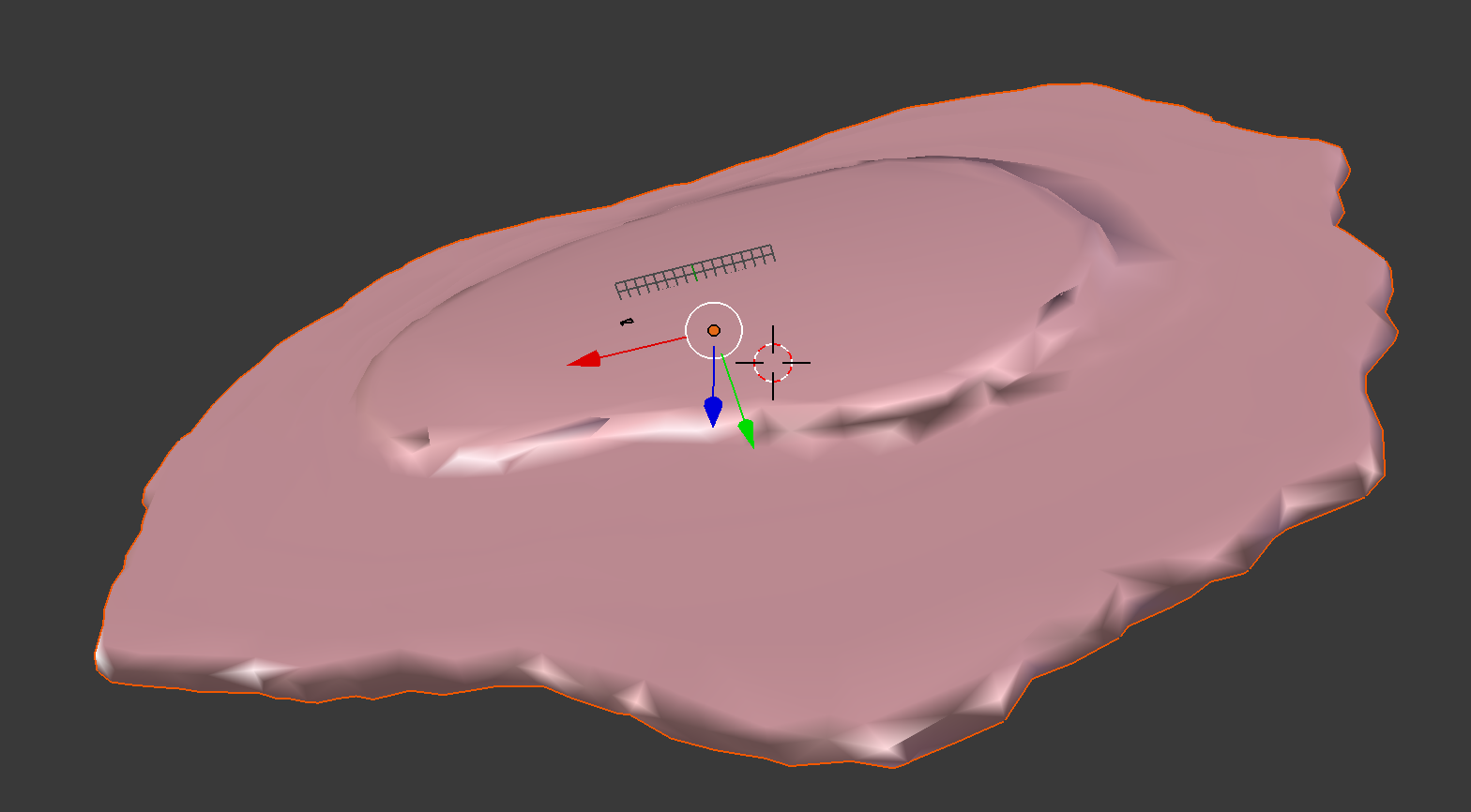
demo3D11¶
Train 3D generative model of the cell framework (nucleus and cell shape) from the entire Murphy Lab 3D HeLa dataset.
demo3D12¶
Train 3D generative model of the nucleus, cell shape, and lysosome from all LAMP2 images in the Murphy Lab 3D HeLa dataset.
demo3D13¶
Export images synthesized by demo3D01 as object files importable to Blender.
demo3D15¶
Synthesize one multichannel 3D image from an endosomal model and diffeomorphic nuclear and cell shape model. The sampling method was set to render endosomes as ellipsoids without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
demo3D16¶
This method shows how to preprocess raw images to use as input for CellOrganizer. The main idea behind this demo is to show the user they can use their own binary images from raw experimental data they can use to synthesize protein patterns. The current demo assumes the resolution of the images is the same as the images that were used to train the protein model
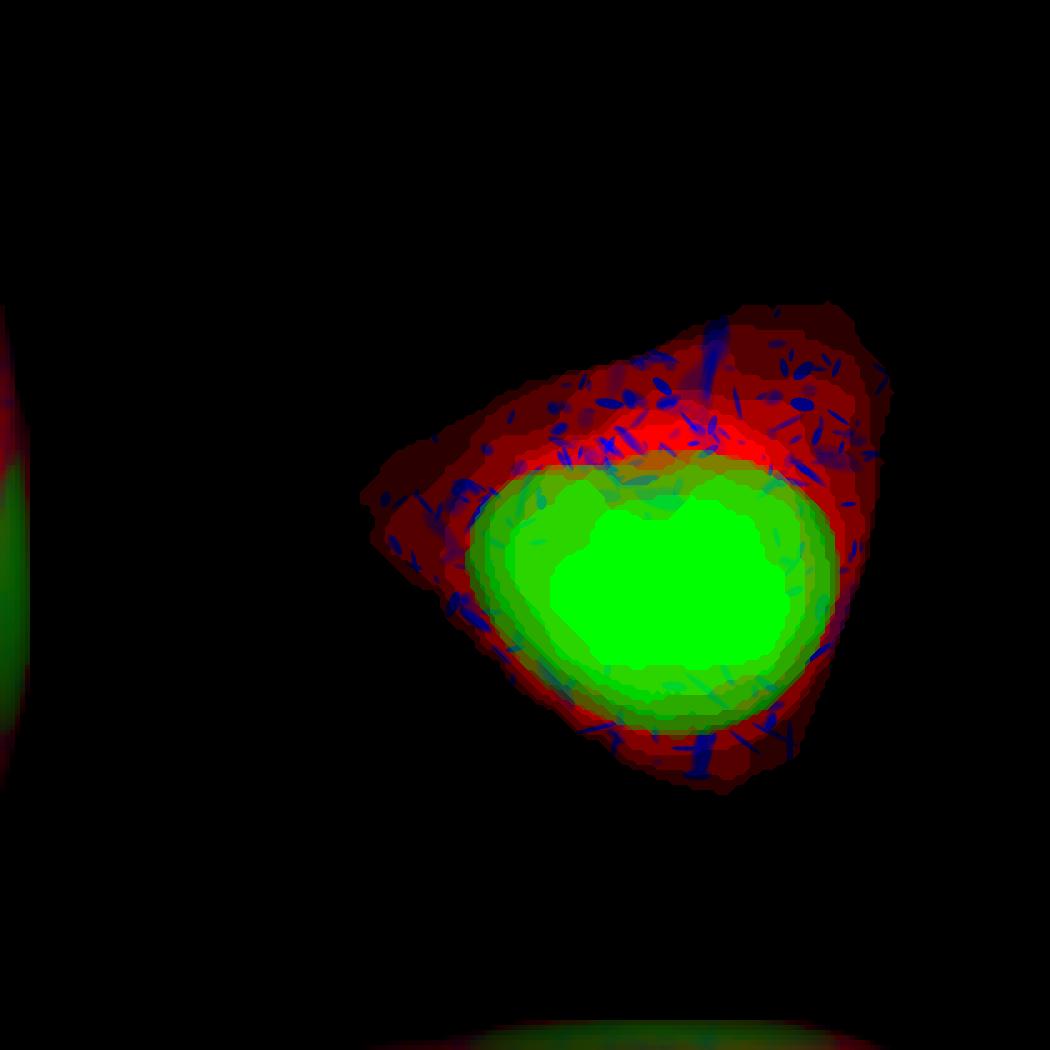
demo3D17¶
The main idea behind this demo is to show the user they can use their own binary images from raw experimental data to synthesize protein patterns.
The current demo assumes the resolution of the images is the same as the images that were used to train the protein model.
demo3D19¶
This demo uses slml2report to compare the parameters between CellOrganzier models.
demo3D20¶
Train 3D generative diffeomorphic nuclear and cell shape model and a lysosomal model from all LAMP2 images in the Murphy Lab 3D HeLa dataset.
demo3D22¶
Synthesizes a protein pattern instance from the synthetic image produced in demo3DDiffeoSynth.
demo3D23¶
Train 3D generative diffeomorphic nuclear and cell shape model and a lysosomal model from all LAMP2 images in the Murphy Lab 3D HeLa dataset.
demo3DSBML¶
This demo converts a sample SBML file to an SBML-spatial instance using the “matchSBML” function. This function takes an SBML file, matches the compartments in the file with available models and synthesizes the appropriate instances.
demo3DMultiresSynth¶
Synthesize multiple 3D images from a lysosome model at different resolutions. This demos show the user can specify the output resolution of the synthesized images.
demo3DObjectAvoidance¶
Synthesizes one image using a lysosomal model with sampling mode set to ‘disc’, no convolution using the object avoidance methods Results will be three TIFF files, one each for cell boundary, nuclear boundary, and lysosomes, in folder “synthesizedImages/cell1” It generates OBJ files that can be imported into Blender.
demo3DPrimitives¶
Synthesizes 1 image using a lysosomal model with sampling mode set to ‘disc’, no convolution and output.SBML set to true Results will be three TIFF files, one each for cell boundary, nuclear boundary, and lysosomes, in folder “synthesizedImages/cell1” Additionally, in the folder “synthesizedImages/” will be a SBML-Spatial(v0.82a) formatted .xml file containing constructed solid geometry(CSG) primitives for lysosomes and parametric objects for the cell and nuclear shapes.
These files can then be read into VCell using the built in importer or CellBlender using the helper function provided in this distribution.
demo3D26¶
This function displays a shape space of some dimensionality. This demo uses the model trained in Johnson 2015.
demo3D27¶
This demo performs a regression between two sets of related shapes (i.e. predicts cell shape from nuclear shape) and displays the residuals as in Figure 2 of Johnson et al 2015.
demo3D28¶
Synthesize one 3D image with nuclear, cell shape, and nucleolar channels from nucleolar model with sampling method set to render nucleoli as ellipsoids without convolution. The model was trained from the Murphy Lab 3D HeLa dataset.
demo3D29¶
This demo shows how an end-user can use experimental data to synthesize a framework.
demo3DDiffeoSynth_uniform¶
This demo illustrates how to sample uniformly at random from a diffeomorphic model.